DATA MED
Data Project : Harnessing the power of data to understand pediatric cancers
Overview
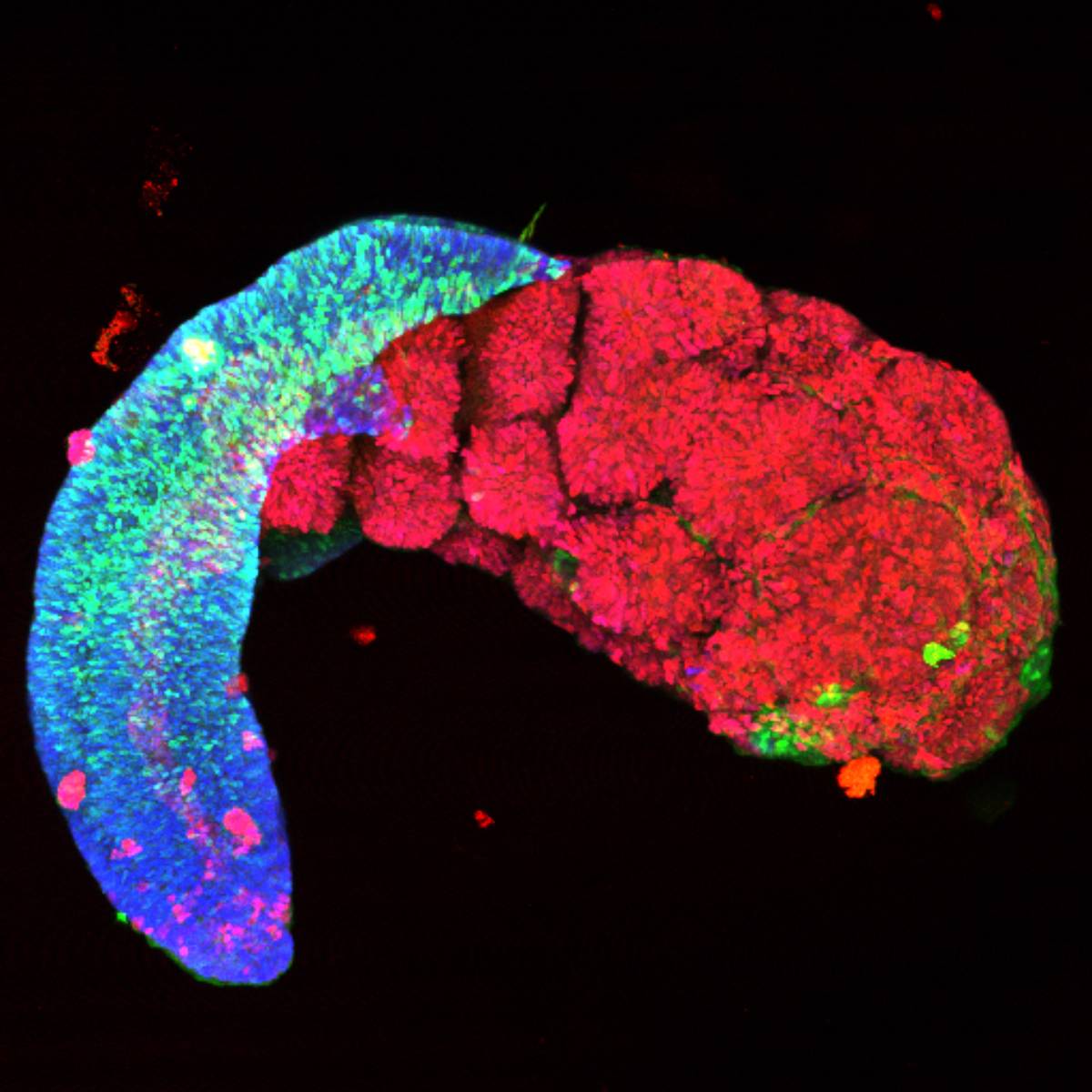
The Cell-ID Data project responds to the challenges posed by the analysis of massive and complex data from innovative technologies. This data, generated at the unicellular level, is essential for deciphering the mechanisms that disrupt cellular destiny and cause the development of pediatric cancers.
Thomas Walter, head of the “Statistical Machine Learning and Modelling of Biological Systems” team of the U1331 “Computational Oncology” unit of the Institut Curie.
Daniel Jost, head of the “Physical Biology of Chromatin” team of the UMR 5239 and INSERM 1293 “Biology and Modelling of the Cell” (LBMC) unit.
Key objectives
- Create a data management infrastructure capable of storing, sharing and securing data while ensuring compliance with legal and ethical constraints.
- Develop analytical tools based on artificial intelligence to extract crucial information from omic data, images and other sources.
- Ensure interoperability with international initiatives to maximize the impact of discoveries and integrate data into large-scale projects.
Resources deployed
Data management infrastructure
- Implementation of a data management plan (DMP) to define the rules for collection, documentation, security and sharing.
- Deployment of a cyberinfrastructure capable of processing petabyte-scale data, relying on local and national networks, such as the bioinformatics platform of the Institut Curie.
- Development of a secure web portal providing centralized access to anonymized and pre-processed data for downstream analysis.
Analysis and integration tools
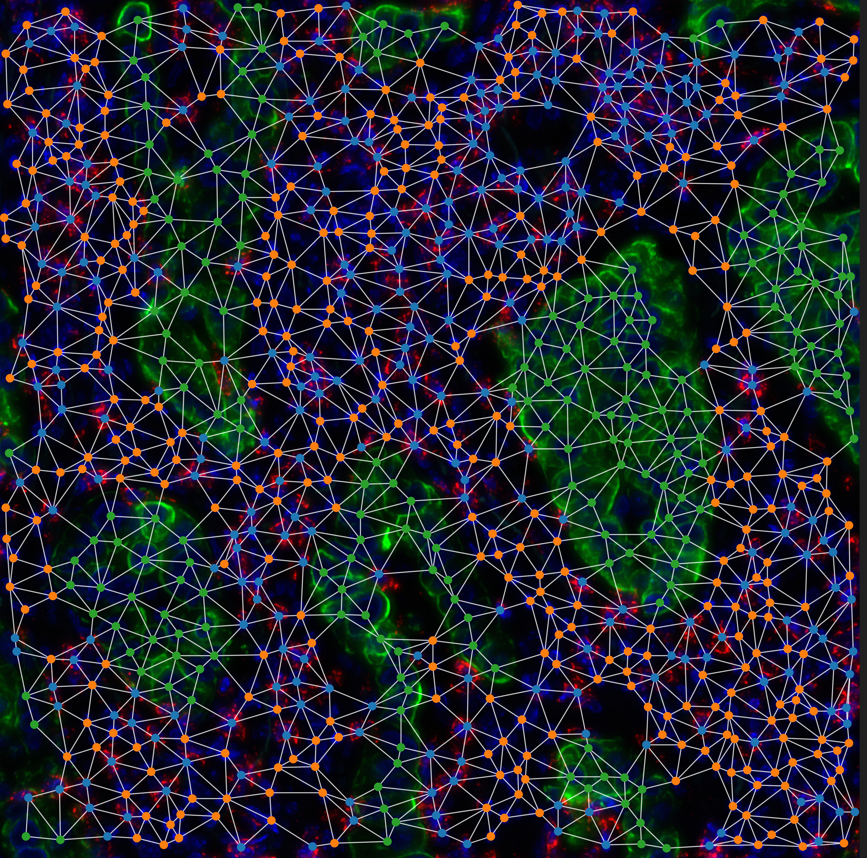
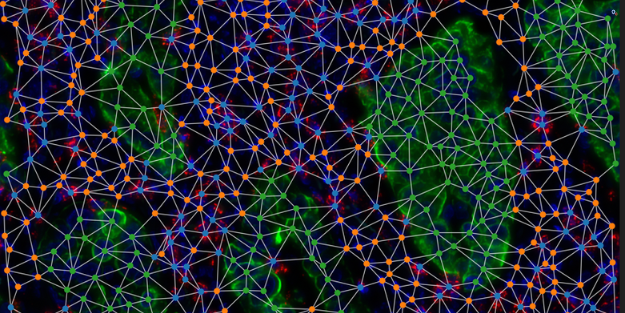
- Creation of bioinformatics pipelines to pre-process and analyze data from technologies such as spatial transcriptomics and mass spectrometry.
- Design of artificial intelligence models capable of imputing missing data, analyzing 2D and 3D images, and linking omic information to cellular phenotypes.
- Development of multimodal integration solutions to combine different data sources and reconstruct cellular trajectories through time and space.
Sharing and dissemination of results
- Publication of the data and tools developed in open access, following the FAIR principles (Findable, Accessible, Interoperable, Reusable).
- Collaboration with international initiatives such as the Human Cell Atlas (HCA) and Euro-BioImaging to ensure the visibility and interoperability of the data produced.
Expected results
The Data project will provide an unprecedented resource for research into pediatric cancers, including:
A centralized database of cellular profiles.
- Pipelines and algorithms accessible to the scientific community.
- Predictive models to identify early abnormalities and propose therapeutic interception strategies.
By combining infrastructure, AI and international collaborations, Cell-ID’s Data project aims to transform data into actionable knowledge to improve the management of pediatric cancers.
Project Med: Intercepting pediatric brain tumors
Overview
The MED project aims to understand and intercept the onset and progression of pediatric brain tumors, such as:
- Medulloblastoma (MB)
- High-grade pediatric gliomas (pHGG), including:
- Diffuse midline gliomas (DMG)
- Hemispheric gliomas (DHG) with histone H3.3
Atypical teratoid rhabdoid tumors (ATRT)
Laure Bally-Cuif, head of the “Neurogenetics of the zebrafish” team of the CNRS UMR3778 “Development biology and stem cells” unit of the Pasteur Institute.
David Castel, researcher in the “Genomics and oncogenesis of pediatric brain tumors” team of the UMR 981 “Molecular predictors and new targets in oncology” of the Gustave Roussy Institute.
Why these particular tumors?
- Complexity of the “original cell”: the same mutation can generate different tumors depending on the original cell type or the microenvironment.
- Common epigenetic alterations: these tumors frequently present mutations affecting chromatin regulation.
- Early development: these cancers often develop during the embryonic period or shortly after birth.
Key objectives
Map the early stages of tumor development
- Study the (epi)genetic signatures and the cells of origin in animal models (mice, drosophila) and brain organoids.
- Identify the events that disrupt cell differentiation and promote tumor transformation.
Validate the hypotheses in human models
- Use brain organoids derived from patient stem cells to recreate and study tumor development.
- Use primary tumor samples to identify specific signatures of cellular deregulation.
Identify “second hits” that promote tumor progression
- Perform CRISPR screening to discover critical epigenetic mutations or alterations.
- Test treatments targeting these alterations in experimental models.
Translate discoveries into clinical interventions.
- Develop strategies to detect and treat tumor recurrence.
- Test “epidrugs” and other therapeutic agents aimed at restoring cell differentiation or slowing tumor progression.
Long-term impact
The results of the MED project will enable:
- Earlier diagnosis through the identification of specific molecular signatures.
- New therapeutic approaches to improve the monitoring and management of pediatric brain tumors.
- Extended applications for other types of pediatric cancers and related neurodevelopmental disorders.
Les autres projets PEPR